Published by Yuri Poluektov on
Please Note: MBL International will be shutting down its operations effective December 31, 2024. Distribution of MBL products in the United States will be transferred to Cosmo Bio US on January 1st while European Distributors will remain unchanged. For any US inquiries regarding orders or support during this transition, reach out to Cosmo Bio: https://www.cosmobiousa.com/. For Non-US inquiries reach out to MBL in Japan: https://www.mblbio.com/.
Published by Yuri Poluektov on Nov 19, 2020 12:00:00 PM

In our previous blog post (How to screen SARS-CoV-2 peptides to facilitate T cell research) we have described how the immune system samples all of the pathogenic proteins by looking at the small fragments of each protein and making a determination on whether that fragment (also referred to as peptide) belongs in the body. We have also described in our previous blog post (Find Human MHC Class I Dominant Peptides) the multiple MHC alleles that we tested for their ability to present SARS-CoV-2 peptides.
In our experiments of determining the ability of A2, A3, A11 and A24 MHC alleles to bind SARS-CoV-2 derived peptides we found little overlap between the peptides presented by human alleles and the H-2Kb mouse allele.
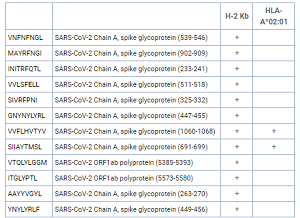
While a purely theoretical exercise at this point, the discovery of peptide sequences that could bind a human allele as well as a mouse allele could become very useful should a SARS-CoV-2 mouse model become available. We have performed an additional study to find SARS-CoV-2 peptides that can be presented by the H-2Kb mouse MHC molecule and 2 of those peptides (VVFLHVTYV and SIIAYTMSL) also proved to be strong binders of the HLA-A*02:01 human allele.
We encourage further trials with our QuickSwitch™ system to help the development of a functional SARS-CoV-2 mouse model that could lead to the creation of a robust COVID-19 vaccine.
| H-2 Kb | HLA-A*02:01 | ||
| VNFNFNGL | SARS-CoV-2 Chain A, spike glycoprotein (539-546) | + | |
| MAYRFNGI | SARS-CoV-2 Chain A, spike glycoprotein (902-909) | + | |
| INITRFQTL | SARS-CoV-2 Chain A, spike glycoprotein (233-241) | + | |
| VVLSFELL | SARS-CoV-2 Chain A, spike glycoprotein (511-518) | + | |
| SIVRFPNI | SARS-CoV-2 Chain A, spike glycoprotein (325-332) | + | |
| GNYNYLYRL | SARS-CoV-2 Chain A, spike glycoprotein (447-455) | + | |
| VVFLHVTYV | SARS-CoV-2 Chain A, spike glycoprotein (1060-1068) | + | + |
| SIIAYTMSL | SARS-CoV-2 Chain A, spike glycoprotein (691-699) | + | + |
| VTQLYLGGM | SARS-CoV-2 ORF1ab polyprotein (5385-5393) | + | |
| ITGLYPTL | SARS-CoV-2 ORF1ab polyprotein (5573-5580) | + | |
| AAYYVGYL | SARS-CoV-2 Chain A, spike glycoprotein (263-270) | + | |
| YNYLYRLF | SARS-CoV-2 Chain A, spike glycoprotein (449-456) | + |
Summary of SARS-CoV-2 peptide binding to mouse H-2 Kb MHC molecules.
Click the link below to view a full description of experiment from the Poluektov, Daftarian and Delcommenne manuscript.
References:
1.Yiwen Zhang et al.The ORF8 Protein of SARS-CoV-2 Mediates Immune Evasion through Potently Downregulating MHC-I bioRxiv 2020.05.24.111823; doi: https://doi.org/10.1101/2020.05.24.111823
2.Chao Hu et al. Identification of Cross-Reactive CD8+ T Cell Receptors with High Functional Avidity to a SARS-CoV-2 Immunodominant Epitope and Its Natural Mutant Variants bioRxiv 2020.11.02.364729; doi: https://doi.org/10.1101/2020.11.02.364729