Published by Yuri Poluektov on Oct 28, 2020 12:00:00 PM
In our previous blog post (How to screen SARS-CoV-2 peptides to facilitate T cell research) we have described how the immune system samples all of the pathogenic proteins by looking at the small fragments of each protein and making a determination on whether that fragment (also referred to as peptide) belongs in the body. Not all peptides are presented equally and most of the amino acids that make up the proteins of the pathogen remain completely invisible to our immune system. One way the immune system can expand its ability to sample a larger variety of pathogen-derived peptide fragments is to have multiple varieties of MHC molecules each geared to detect a certain type of peptide.
Every human has 3 MHC Class I genes and 3 MHC Class II genes on Chromosome 6 responsible for the production of 3 Class I and 3 Class II molecules that present the pathogenic peptide fragments to the immune system. Since there are 2 copies of each chromosome, a human, under normal conditions, can have up to 6 MHC Class I and 6 MHC Class II alleles, provided that there are no duplicates in the genes. Each commonly occurring human MHC molecule discovered is assigned a name starting with the “HLA-“designation. The complete makeup of an individual’s 12 MHC molecules is known as the MHC Haplotype. Some MHC types are very common and occur in many individuals throughout the world, while others are much less so, but each persistent MHC type has provided some evolutionary advantage in the past.
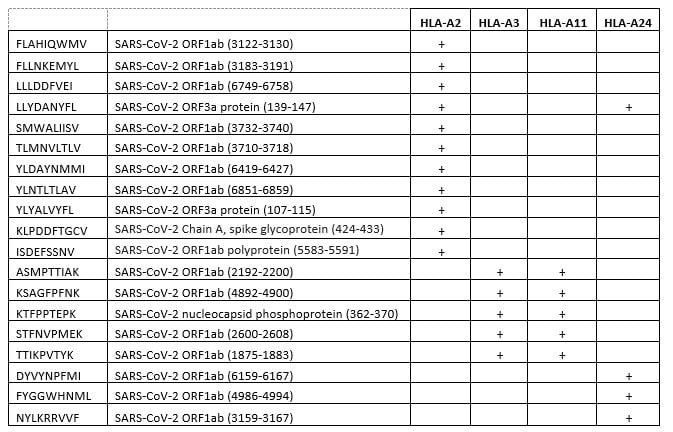
While designing our QuickSwitch™ platform at MBL International, we examined the available MHC Haplotypes and tried to develop a QuickSwitch™ system for the most widely studied MHC molecules. In addition to the HLA-A*02:01 (A2) molecule we also examined the binding of SARS-CoV-2 peptides to HLA-A*03:01 (A3), HLA-A*11:01 (A11) and HLA-A*24:02 (A24) molecules. While some peptides overlapped in their binding between different MHC alleles, like in the case of HLA-A3 and HLA-A11, which are structurally similar, most of the SARS-CoV-2 peptides we tested only bind one of the MHC alleles.
While our results do not encompass the entire SARS-CoV-2 proteome, we hope that our QuickSwitch™ platform will be used by researchers in the future to test if a viral peptide, derived from a critical viral structure that they are studying, can be presented to the immune system by one of the MHC alleles available in our platform.
| HLA-A2 | HLA-A3 | HLA-A11 | HLA-A24 | ||
| FLAHIQWMV | SARS-CoV-2 ORF1ab (3122-3130) | + | |||
| FLLNKEMYL | SARS-CoV-2 ORF1ab (3183-3191) | + | |||
| LLLDDFVEI | SARS-CoV-2 ORF1ab (6749-6758) | + | |||
| LLYDANYFL | SARS-CoV-2 ORF3a protein (139-147) | + | + | ||
| SMWALIISV | SARS-CoV-2 ORF1ab (3732-3740) | + | |||
| TLMNVLTLV | SARS-CoV-2 ORF1ab (3710-3718) | + | |||
| YLDAYNMMI | SARS-CoV-2 ORF1ab (6419-6427) | + | |||
| YLNTLTAV | SARS-CoV-2 ORF1ab (6851-6859) | + | |||
| YLYALVYFL | SARS-CoV-2 ORF3a protein (107-115) | + | |||
| KLPDDFTGCV | SARS-CoV-2 Chain A, spike glycoprotein (424-433) | + | |||
| ISDEFSSNV | SARS-CoV-2 ORF1ab polyprotein (5583-5591) | + | |||
| ASMPTTIAK | SARS-CoV-2 ORF1ab (2192-2200) | + | + | ||
| KSAGFPFNK | SARS-CoV-2 ORF1ab (4892-4900) | + | + | ||
| KTFPPTEPK | SARS-CoV-2 nucleocapsid phosphoprotein (362-370) | + | + | ||
| STFNVPMEK | SARS-CoV-2 ORF1ab (2600-2608) | + | + | ||
| TTIKPVTYK | SARS-CoV-2 ORF1ab (1875-1883) | + | + | ||
| DYVYNPFMI | SARS-CoV-2 ORF1ab (6159-6167) | + | |||
| FYGGWHNML | SARS-CoV-2 ORF1ab (4986-4994) | + | |||
| NYLKRRVVF | SARS-CoV-2 ORF1ab (3159-3167) | + |
Summary of SARS-CoV-2 peptide binding to human MHC class I molecules.
