Published by Yuri Poluektov on Aug 17, 2020 12:00:00 PM
Whenever our immune system is faced with a challenge, be it a multicellular parasite, a bacteria, a virus, or even cancer, this threat is detected and sampled primarily by the protein sequence of the pathogen in question. Other aspects of the pathogen, such as its glycosylation pattern or the peculiar structures of its RNA and DNA molecules could have a strong effect on how the immune system deals with it. But, overall, the adaptive aspect of the mammalian immune system has evolved over many years to examine the protein sequences that make up the invading pathogen. The key function of our immune system relies on its ability to constantly sample all proteins present in our body and determine whether it is a protein that belongs or a protein that must be eradicated together with any cells or organisms that produce it. Unfortunately, most proteins are too large for the immune system to sample at once and so it has developed a mechanism to cut the full-length proteins into small stretches of amino acids which are examined individually.
While many assume that this cutting and sampling process will eventually be able to probe every amino acid of every protein present in our body, in reality, this process is less than perfect and not all stretches of the protein sequence are processed by the immune system. In some cases, it is because the pathogen has developed a means to elude the immune system, but in many cases, it is simply because the cut peptide pieces are not able to bind the molecules that present them to the immune system, commonly known as the MHC molecules.
The ongoing COVID-19 pandemic has put a lot of pressure on developing a SARS-CoV-2 vaccine. But, just like with any other pathogen, the amino acid sequence of the proteins that the SARS-CoV-2 virus is made up of is not all equally presented to the immune system. Some peptides are great binders of the MHC molecules while others are not. To help the global search for a vaccine we used the QuickSwitch™ MHC Tetramer platform to determine some of the SARS-CoV-2 peptides that are well presented to the human immune system by human Class I and II MHC molecules.
The QuickSwitch™ platform produced and sold by MBL International is primarily a means to detect T cells stimulated in the immune system by a specific pathogen-derived peptide. It contains a recombinant form of either Class I or Class II MHC molecules designed in such a way that it can be loaded with any peptide of your choice to sample any T cell specific for it, but only if that peptide is a strong binder of the MHC molecule and hence is readily detectable by the immune system. By focusing on this aspect of the QuickSwitch™ platform we were able to screen a number of peptides derived from some of the SARS-CoV-2 proteins of interest and determine their ability to replace, or exchange, the exiting (QuickSwitch™) peptide present in the MHC molecule.
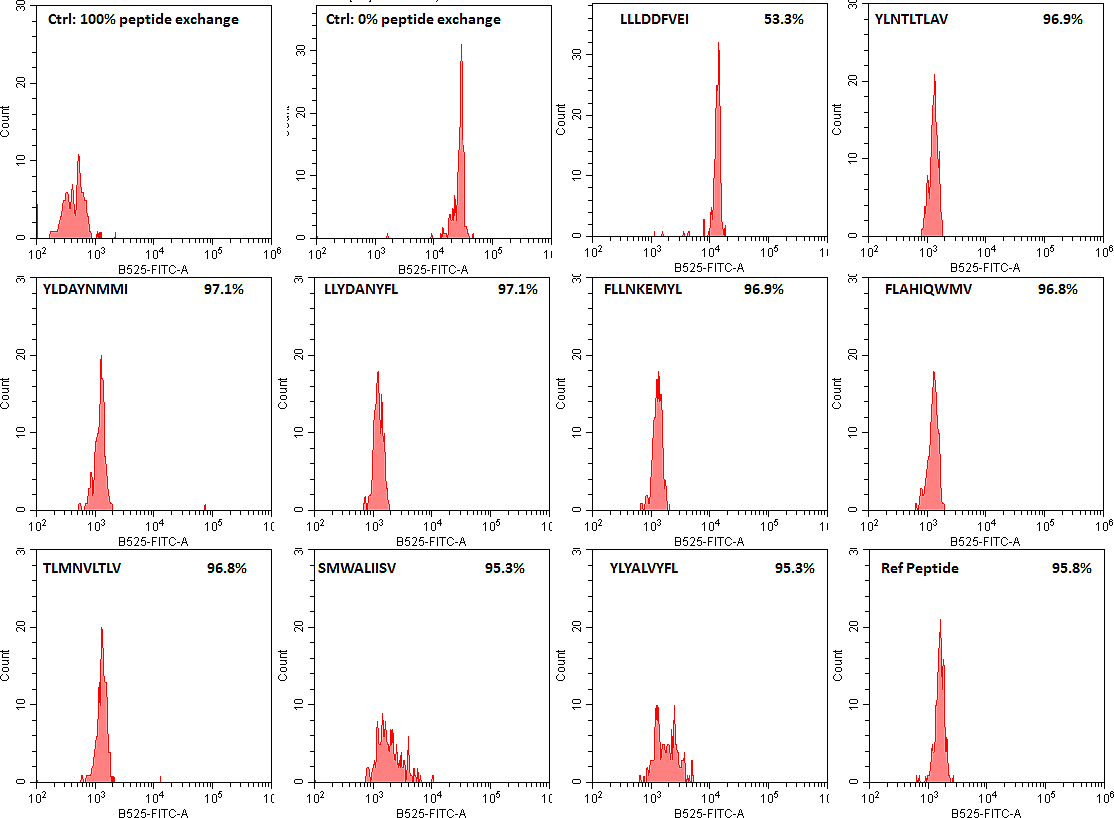
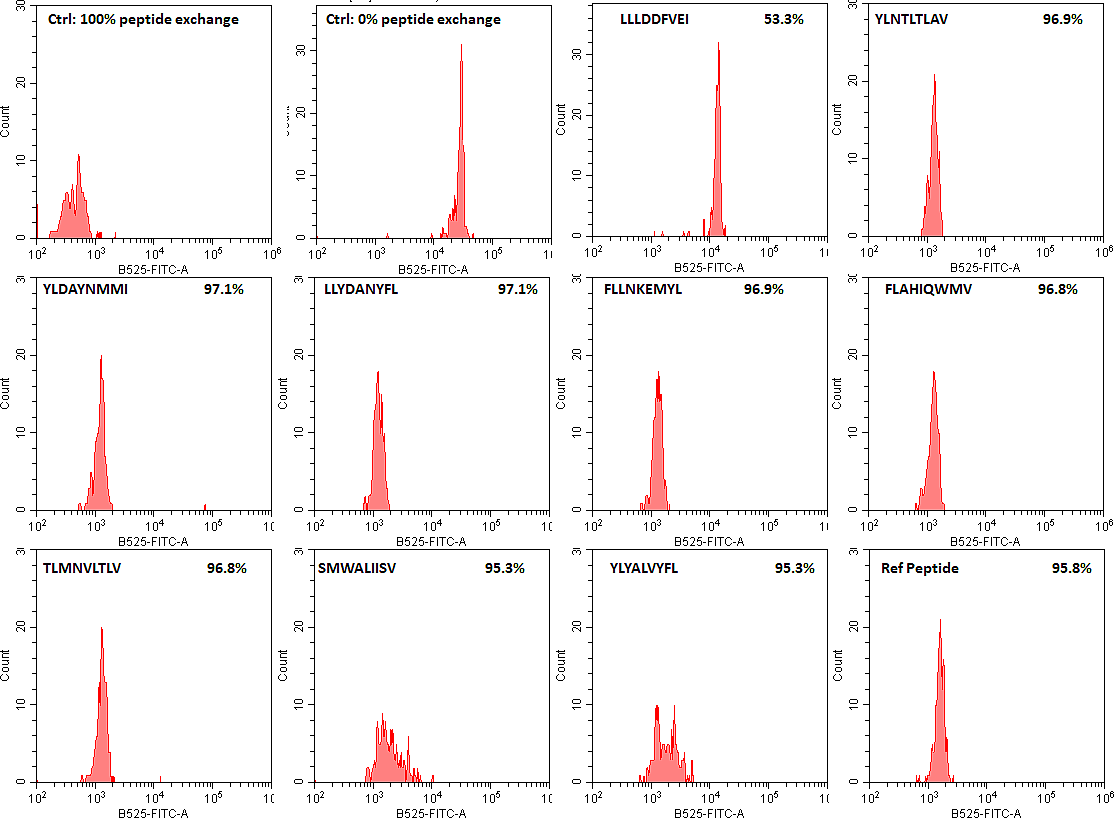
This exchange can be visualized by binding up the QuickSwitch™ tetramer molecules with magnetic beads and staining the bound MHC molecules with a FITC-labeled antibody specific for the exiting QuickSwitch™ peptide (both components provided in the kit). The magnetic beads are designed to be easily detectable by all flow cytometers and the Mean Fluorescence Intensity (MFI) of the detected FITC signal is representative of the amount of QuickSwitch™ peptide remaining bound to the MHC molecule following the peptide exchange procedures. If no competing peptide is added to the exchange reaction, then the resulting FITC signal will be high. If a high-binding reference peptide (provided in the QuickSwitch™ kit for each available MHC allele) is added to the exchange reaction then it will replace all of the QuickSwitch™ exiting peptide resulting in very little detected FITC signal. Using the non-exchanged control, a percentage of peptide exchange can be estimated for each SARS-CoV-2 peptide tested, if at least 90% of the exiting peptide is exchanged, it is a good indication that the viral peptide can be presented to the human immune system.
Our screen of SARS-CoV-2 peptides has determined a number of good MHC class I and II binders as well as some peptides that are not (flow cytometry data of HLA-A*02:02 binders are shown as examples). We hope that this will facilitate future COVID-19 vaccine research by limiting the T cell studies to peptides that can be presented by the immune system and not focusing on the peptides that our immune system cannot process.
SARS-CoV-2 peptide exchange with QuickSwitch exiting peptide loaded HLA-A*02:01 Tetramer-PE molecules.
Click the link below to view a full description of experiment from the Poluektov, Daftarian and Delcommenne manuscript.
