Tags and Tag antibodies
What are epitope tags?
Antibodies are an essential tool to determine the localization and expression of target proteins, and for purification in the presence of numerous other proteins in living organisms. However, specific antibodies to a target protein are not always available. In that case, the target protein can be added a marker (tag) by genetic engineering techniques and expressed in the living organism. A specific antibody to the tag can distinguish the target protein from other similar proteins.
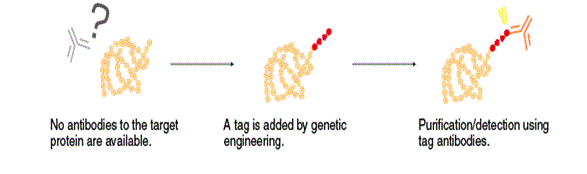
.png?width=352&height=243&name=Capture-1%20(1).png) The tags are relatively small polypeptides, ranging from a few to several tens of amino acids in length. Antibodies that recognize a tag are commonly called tag antibodies, and the tag sequence that serves as an epitope for tag antibodies is called an “epitope tag”. Many different types of epitope tags have been developed for different purposes. Among them, the FLAG® (DYKDDDDK, DDDDK, etc.), HA, His, Myc, and V5 tags are commonly used because expression cassette vectors for these tags are widely available. Other types of tags include fluorescent proteins, such as GFP and RFP, and special purpose proteins, such as GST and MBP. These tags are used for specific purposes.
The tags are relatively small polypeptides, ranging from a few to several tens of amino acids in length. Antibodies that recognize a tag are commonly called tag antibodies, and the tag sequence that serves as an epitope for tag antibodies is called an “epitope tag”. Many different types of epitope tags have been developed for different purposes. Among them, the FLAG® (DYKDDDDK, DDDDK, etc.), HA, His, Myc, and V5 tags are commonly used because expression cassette vectors for these tags are widely available. Other types of tags include fluorescent proteins, such as GFP and RFP, and special purpose proteins, such as GST and MBP. These tags are used for specific purposes.
*FLAG® is a trademark of Sigma-Aldrich Co. LLC.
Type of epitope tags
- Peptide tags
- Polypeptide tags
- Tags for the AID system
- Luciferase
- Fluorescent proteins
- Haptens
Peptide tags
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
DDDK (FLAG®)
|
Artificial Sequence
|
8 amino acids (1.0 kDa) (DYKDDDDK and its variants)
|
Well known as the FLAG® tag (SIGMA
|
|
|
HA
|
Virus
|
9 amino acids (1.1kDa) (YPYDVPDYA)
|
Identified as the antibody recognition site within the envelope glycoprotein of influenza virus, hemagglutinin. (Wilson IA et al. PMID: 6204768)
|
|
|
6xHis
|
Artificial sequence
|
6 amino acids (0.8 kDa) (HHHHHH)
|
Can be used for protein purification using metal-chelating columns, such as a nickel column. This technique is suitable for low-cost, large-scale protein purification. Nickel columns can be used in purification of proteins denatured by urea or guanidine hydrochloride.
|
|
|
Myc
|
Human c-Myc protein
|
10 Amino acids (1.2 kDa) (EQKLISEEDL)
|
c-Myc protein is a transcription factor involved in the cell cycle and apoptosis. Abnormalities in c-Myc, such as mutations and overexpression, are associated with various cancers of the hematopoietic system.
|
|
|
V5
|
Virus
|
14 Amino acids (1.4 kDa)(GKPIPNPLLGLDST)
|
This peptide sequence is present in the P and V proteins of Simian virus 5 (SV5) of the Rubulavirus genus in the Paramyxoviridae family.
|
|
|
S
|
Human pancreatic RNASE A
|
15 Amino acids (1.7 kDa) (KETAAAKFERQHMDS)
|
The S-tag tightly binds to S-protein to form RNase S. Using this feature, activity-based detection and quantitative analysis can be performed.
|
|
|
E
|
Osteocalcin fragment
|
13 Amino acids (1.4 kDa) (GAPVPYPDPLEPR)
|
This peptide sequence is present in a bone hormone osteocalcin produced by osteoblasts.
|
|
|
T7
|
Virus
|
11 Amino acids (1.2 kDa) (MASMTGQQMG)
|
N-terminal peptide of the capsid protein of T7 phage.
|
|
|
VSV-G
|
Virus
|
11 Amino acids (1.3 kDa) (YTDIEMNRLGK)
|
“VSV-G” is an acronym of vesicular stomatitis virus G glycoprotein. This peptide sequence is present in the virus envelope protein.
|
|
|
Glu-Glu
|
Virus
|
9 Amino acids (1.2 kDa) (EEEEYMPME)
|
Polyomavirus middle T antigen
|
|
|
Strep-tag II
|
Artificial sequence
|
8 Amino acids (1.1 kDa) (WSHPQFEK)
|
Can bind to the biotin-binding site of streptavidin. Protein purification systems using the biotin-streptavidin interaction are available.
|
|
|
HSV
|
Virus
|
11 Amino acids (1.2 kDa) (QPELAPEDPED)
|
The peptiide sequence is present in the envelope protein of herpes simplex virus.
|
|
|
CBD (Chitin Binding Domain)
|
E. Coli
|
29 Amino acids (3.1 kDa) TTNPGVSAWQVNTAYTAGQLVIYNGKTYK
|
Commonly used for protein purification. A target protein is fused to the intein and the chitin binding domain (CDB). Upon induction of self-cleavage of the intein, the target protein alone is isolated.
|
|
|
CBD (Calmodulin Binding Peptide)
|
Rabbit
|
26 Amino acids (3.0 kDa) (KRRWKKNFIAVSAANRFKKISSSGAL)
|
Calmodulin binding protein (CBP) tightly binds to calmodulin in the presence of calcium. This feature is used for calmodulin-affinity chromatography under harsh washing conditions to minimize non-specific binding. Because E-coli proteins do not interact with calmodulin, recombinant proteins can be obtained at high purity.
|
Polypeptide tags
Polypeptide tags enhance the solubility of expressed proteins. Recombinant proteins that tend to become insoluble in E. coli can be expressed as soluble proteins when fused to these tags.
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
GST (Glutathione-S-transferase)
|
Parasite (Schistosoma japonicum)
|
26 kDa
|
GST fusion proteins can be affinity-purified using interaction with the substrate of GST, glutathione. Because GST could interfere with protein folding and function due to its relatively high molecular weight, it is desirable to cleave the tag after purification.
|
|
|
MBP (Maltose Binding Protein)
|
E. Coli
|
43 kDa
|
MBP fusion proteins can be purfied using an amylose column. MBP fusion proteins captured by the column are eluted with maltose due to their affinity for the sugar.
|
|
|
Thioredoxin (Trx)
|
E. Coli
|
12 kDa
|
Thioredoxin is a protein involved in various oxidation-reduction reactions in the living organisms. Human cytokines and growth factors often form inclusion bodies when expressed in E. coli. Thioredoxin fusions of these proteins have higher solubility and can be purified in their active form.
|
Tag for the AID system
These tags are used for the AID system, which is a tunable protein expression system using a plant hormone. In this system, proteins expressed in animal cells can be rapidly degraded on command.
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
mini-AID
|
Plant hormone
|
Full-length AID-tag (IAA17): 229 amino acids; mini-AID tag: 68 amino acids
|
These tags are used in the AID system. In this system, a recombinant protein expressed in the cell can be rapidly degraded by the addition of the plant hormone auxin. (Nishimura K et al. PMID: 19915560)
|
Reporter genes
Luciferase (luminescent enzyme)
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
Luciferase
|
Firefly
|
61 kDa
|
Luciferase is a luminescent enzyme isolated from the firefly. Luminescence is generated when the enzyme reacts with the substrate luciferin and ATP in the presence of magnesium. Yellow-green light (emission maximum at 562 nm) is emitted. Luciferase is commonly used as a reporter gene for the analysis of transcription activity of the promoter/enhancer region of a gene.
|
|
|
Renilla Luciferase
|
Sea pansy
|
36 kDa
|
Upon activation by calcium binding, Renilla luciferase catalyzes the oxidation of the luminescent substrate Renilla luciferin. Blue-green light (emission maximum at 492 nm) is emitted.
|
Fluorescent proteins
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
GFP
|
Jellyfish
|
27 kDa
|
GFP fusion proteins can be expressed and detected by fluorescence. Anti-GFP antibodies are used for higher sensitivity and for detection of non-fluorescent (denatured) fusion proteins in samples, such as formalin-fixed, stained samples and Western blot membranes
|
|
|
Renilla GFP
|
Sea pansy
|
26 kDa
|
Renilla GFP is as bright as EGFP, but its fluorescence fades more slowly. Renilla GFP is also less cytotoxic than EGFP.
|
|
|
RFP
|
Variant of coral-derived DsRed
|
27 kDa
|
RFP is a variant of the fluorescent protein DsRed isolated from the coral Discosoma sp. While DsRed forms a tetramer, many variants that emit fluorescence as monomers have been developed.
|
Others
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
β-galactosidase
|
E. Coli
|
116 kDa
|
β-Galactosidase is a protein encoded by the E. coli lacZ gene. This enzyme is necessary for the degradation of lactose to glucose to obtain energy through glycolysis. β-Galactosidase is well-known in the area of genetic engineering because its chromogenic reaction with the substrate X-Gal is widely used as an indicator of the presence of a recombinant gene.
|
Haptens
| TAG | Origin | SIZE (PEPTIDE SEQUENCE) | FEATURES | MBLI PRODUCT PAGE |
|---|---|---|---|---|
|
Digoxigenin (DIG)
|
Plant-derived steroid
|
390 Da
|
The DIG tag is a hapten that can be easily conjugated to proteins and nucleic acid. This tag is used in various biotechnology applications in a similar manner as biotin and fluorescein. Notably, DIG-conjugated UTP can be used for labeling nucleic acid probes. Used in conjunction with DIG antibodies, DIG-labeled probes made possible non-radioactive Northern blotting, Southern blotting, and in situ hybridization.
|
|
|
FITC
|
Small-molecule fluorescent dye
|
390 Da
|
FITC (fluorescein isothiocyanate) is not simply a fluorescent dye; like biotin and DIG, it is used for labeling proteins and nucleic acids. FITC-labeled proteins and nucleic acids can be directly observed, but its fluorescence tends to fade rapidly. Using an anti-FITC antibody and a labeled secondary antibody provides a higher sensitivity of detection.
|
How to use Tag antibodies
| Method | PURPOSE | |
|---|---|---|
|
Detection
|
Western blottingImmunoprecipitation (IP), Co-immunoprecipitation (Co-IP)Immunostaining (IC, IH)Flow cytometry (FCM)
|
Confirmation of the presence or absence of expressionAnalysis of factors that associate with a tagged protein (Co-IP)Intracellular localization
|
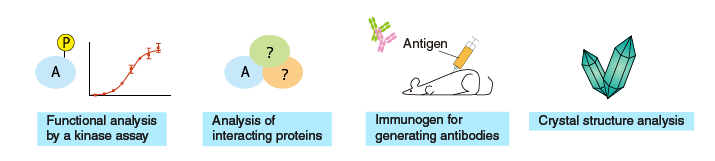
|
Purification
|
Purification using antigen-antibody interaction
|
Analysis of interacting proteins (pull-down assay)Crystal structure analysisImmunogen preparation for generating antibodiesKinase assay
|
Detection methods
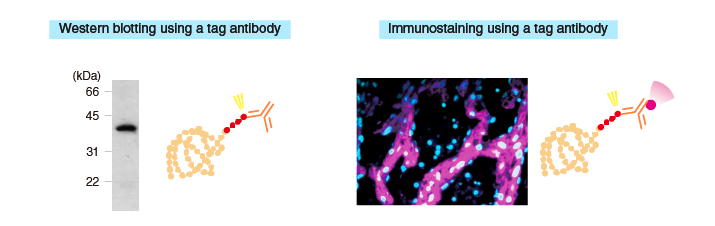
Uses of purified proteins
Advantages and disadvantages of using a tag and tag antibody
|
Advantages
|
Because most tag sequences are not derived from animal cells, tag antibodies are often highly reactive with little non-specific binding.
|
|
|
When good antibodies are not available for a target protein, the protein can be expressed in the cell as a tagged protein and studied for its behavior.
|
|
|
A tag antibody can be used for the detection and purification of multiple proteins with the same tag.
 |
|
|
Some tags can be removed from a tagged protein after purification by a specific protease.
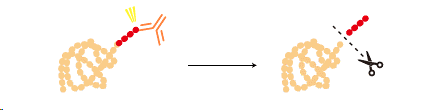
|
|
|
A protein can be fused to two different tags and purified by a two-step process for a higher purity.
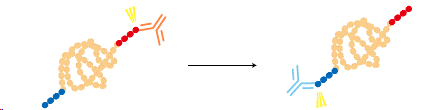
|
|
|
In co-immunoprecipitation, target protein-specific antibodies can interfere with the interaction of the target protein with other proteins. This can be avoided by using a tag and a tag antibody.
|
|
|
An insoluble protein can sometimes be made soluble by the addition of a tag.
|
|
|
Proteins fused with a fluorescent protein can be studied for intracellular localization in living cells.
|
|
Disadvantages
|
Expressed tagged proteins are artificial molecules and may not behave the same way as the endogenous proteins in actual biological reactions.
|
|
|
Depending on the site of the tag (N-terminus, C-terminus, etc.), protein conformation could be affected, causing loss of activity.
|
|
|
When multiple tagged proteins are used in an experimental system, the proteins must be engineered such that each protein has a distinct tag.
|
Related Links
*Antibody
→Antibody basics
→Antibodies as a research tool
- How to select antibodies
- Labeled antibodies
- How to label antibodies
- Main causes of non-specific reactions
- How to reduce non-specific reactions
- Tags and Tag antibodies
*Qualitative and quantitative measurements of proteins using antibodies
- Western blotting (WB)
- Enzyme-linked Immunosorbent assay (ELISA)
- Immunoprecipitation (IP)
- Co-immunoprecipitation (Co-IP)